Data formats and file types: Multidimensional array data
Better Code, Better Science: Chapter 7, Part 5
This is a possible section from the open-source living textbook Better Code, Better Science, which is being released in sections on Substack. The entire book can be accessed here and the Github repository is here. This material is released under CC-BY-NC.
Many forms of data are represented as multidimensional arrays. For example, many forms of microscopy data are represented as either two-dimensional (slice) or three-dimensional (volume) arrays, while dynamic imaging modalities are often represented as three- or four-dimensional data, with the first two or three dimensions representing space and the final dimension reflecting time. Within Python these are generally processed using numpy, which efficiently processes large multidimensional arrays. As an example, I loaded a four-dimensional brain template, where the first three dimensions are spatial and the final dimension reflects 512 different components that define a probabilistic atlas of the brain. After loading the data into a numpy array, we can see its shape:
% data.shape
(91, 109, 91, 512)In this case, the first three dimensions are spatial (representing the left/right, front/back, and up/down axes of the brain) and the final dimension represents timepoints. We can also use the imshow function from matplotlib.pyplot to view a two-dimensional slice of the image at a particular timepoint:
import matplotlib.pyplot as plt
# view z==50 at timepoint 5
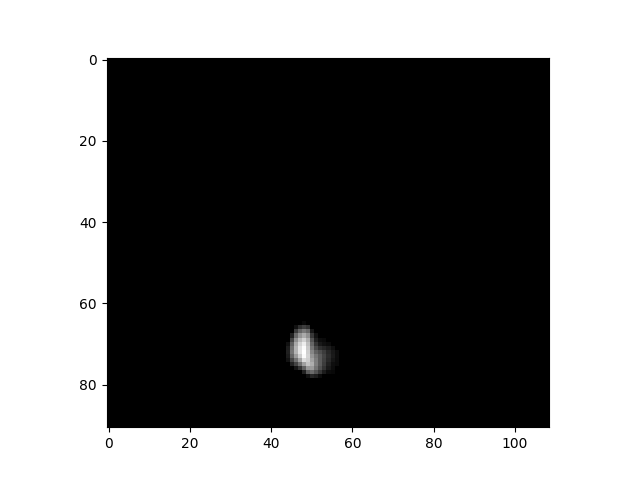
plt.imshow(data[:, :, 50, 5], cmap=’gray’)An example of a slice from a 4d image dataset.
Multidimensional array file formats
A common file format for multidimensional array data in Python is the native Numpy format, which is known by its extension, .npy. This file format has a number of benefits for Python users:
It is very fast to both read and write files
It supports *memory mapping*, which allows portions of the file to be read without loading the entire file
It has perfect fidelity and maintains exact data types
It is very easy to use (
np.load()to load andnp.save()to save).
However, the .npy format also has a number of drawbacks for scientific data:
It is harder for non-Numpy users to work with, requiring specialized libraries to read in languages other than Python.
It does not compress the data, so files can be much larger than a compressed file format. This can become very important when working with large sparse datasets.
It doesn’t support storage of metadata alongside the data
There are several other commonly used standard file formats for array data; we will focus on HDF5 and Zarr. HDF5 is a longstanding format for storage of large datasets, which is supported by nearly all programming languages. It has built-in support for compression, and allows access to specific chunks without loading the entire dataset. However, may researchers (at least within the Python ecosystem) are moving towards the Zarr format, which stores data in a set of smaller chunk files rather than a single large file as in HDF5. Zarr has several advantages over HDF5:
Zarr is much more efficient for cloud storage, since only the specific chunk file needs to be accessed
Zarr is simpler than HDF5, and has a more Pythonic interface
Zarr makes it very easy to add new data to a file, which can be more difficult in HDF5
Let’s use the data from above as an example. This is a dataset that is highly sparse, with most of the values being zero; after thresholding to remove very small values, only about 0.1% of the values are nonzero. This means that we should be able to get a high degree of compression for this dataset, given the redundancy of the data. Let’s first save the data to a .npy file and look at its (uncompressed) size:
np.save(’/tmp/difumo.npy’, data)> du -sm /tmp/difumo.npy
3526 /tmp/difumo.npyThat’s about 3.5 gigabytes. Now let’s save it to HDF5:
import h5py
with h5py.File(’/tmp/difumo.h5’, ‘w’) as f:
f.create_dataset(’difumo’, data=data, compression=’gzip’)> du -sm /tmp/difumo.h5
10 /tmp/difumo.h5Due to the compression by HDF5, the data file is about 350 times smaller with HDF5 than with the Numpy standard format! We can also look at the same with Zarr:
import zarr
zarr_data = zarr.open(’/tmp/difumo.zarr’, mode=’w’, shape=data.shape, dtype=data.dtype)
zarr_data[:] = data> du -sm /tmp/difumo.zarr
5 /tmp/difumo.zarrThis shows that Zarr obtains even more compression than HDF5, each using its default compression method; note that it might be possible to get better compression using custom compression methods for each. We can also compare the time needed to load each of the files; because of the relatively longer loading time for these files, we only perform 10 repetitions in this example:
nreps = 10
for ext in [’npy’, ‘h5’, ‘zarr’]:
start_time = time.time()
filename = f’/tmp/difumo.{ext}’
if ext == ‘npy’:
for _ in range(nreps):
data_loaded = np.load(filename)
elif ext == ‘h5’:
for _ in range(nreps):
with h5py.File(filename, ‘r’) as f:
data_loaded = f[’difumo’][:]
elif ext == ‘zarr’:
for _ in range(nreps):
zarr_data = zarr.open(filename, mode=’r’)
data_loaded = zarr_data[:]
end_time = time.time()
avg_load_time = (end_time - start_time) / nreps
print(f”Average loading time for {ext}: {avg_load_time:.6f} seconds”)Average loading time for npy: 0.451627 seconds
Average loading time for h5: 3.138907 seconds
Average loading time for zarr: 0.745648 secondsThis shows that Zarr is slightly slower than the native numpy file loader, but much faster than HDF5.
I would suggest that unless you have a strong need for HDF5 compatibility, that you use Zarr files to store large binary data files such as matrices.
Symmetrical matrices
It’s not uncommon to work with symmetrical matrices, such as correlation matrices. Since the upper triangle and lower triangle of a symmetric matrix are simply transposed versions of one another, there is no need to save both - we can simply save either the upper or lower triangle. Depending on the application, this may be a useful way to save space and loading time.
In the next post I will move to disussing other types of data, including network/graph data.